Research
Research Statement
My research is centered at the intersection of Deep Learning, Signal Processing, and Computer Vision, with a primary focus on developing ML-based solutions for Healthcare and Diagnostics. I am driven by the challenge of integrating information from multiple domains, signal representations, or modalities to build models that are efficient, generalizable, and interpretable.
In my undergraduate engineering studies, Signal Processing was something I really started to enjoy. To me, the frequency domain was a whole new universe, and that fascination is still intact. Later, I got to explore Machine Learning, and soon I knew I had found my core interest. To bridge these two worlds, I consciously chose to enroll in two simultaneous MS programs (in EE and CS), allowing me to solidify my understanding of core DL and also explore its applications in Signal and Image Processing.
Due to this multidisciplinary background, I have quite a wide range of research interests, spanning from Multi-Modal Machine Learning & Deep Learning Architectures, to their applications in Biomedical Imaging, Signal Processing, Bioinformatics and even Computational Social Science. My research (currently) focuses on effectively fusing features learned from two complementary domains: the time/spatial domain (like a raw audio waveform or an image) and the frequency/spectral domain (its Fourier transform or spectrogram). The goal is to build models that learn from both domains and, as a result, are more efficient and robust, gaining a richer, more holistic view of the data. I've actively explored this "dual-domain" approach in several areas, including Biomedical Imaging (fusing images with their FFTs) and Audio Analysis (combining raw waveforms with spectrograms).
I enjoy designing and extensively tweaking Deep Learning architectures, and following theoretically grounded intuitions while doing so (even though intuition doesn't always work with neural networks!). I am also interested in more theoretical aspects of Deep Learning, like Generalization, and worked on it for my MS thesis. As someone who is passionate about politics and social justice, another interest of mine is applying computational methods to analyze complex socio-political systems. I have already worked on a quantitative analysis of the July Revolution, a pivotal event in Bangladesh’s history that I have followed intensely and have been a part of.
I recently co-founded BIOML (Bioinformatics, Imaging & Omics using Machine Learning), a collaborative of young researchers from Bangladesh. Some of our current projects include developing advanced methods for Antimicrobial Resistance (AMR) prediction, creating interpretable approaches for physiological signal-based diagnostics, and designing novel deep learning frameworks for medical imaging and audio analysis. The ultimate goal is to build intelligent tools that have a tangible impact on real-world challenges.
If any of this sounds interesting, or if you'd like to collaborate, please feel free to send me an email!
Research Interests
- Multi-Modal Machine Learning
- Deep Learning Architectures
- Signal & Image Processing
- Audio and Speech Processing
- Federated Learning
- Computer Vision and Medical Imaging
- Bioinformatics
- AI in Healthcare
- Computational Social Science
- Explainable AI
Thesis Projects
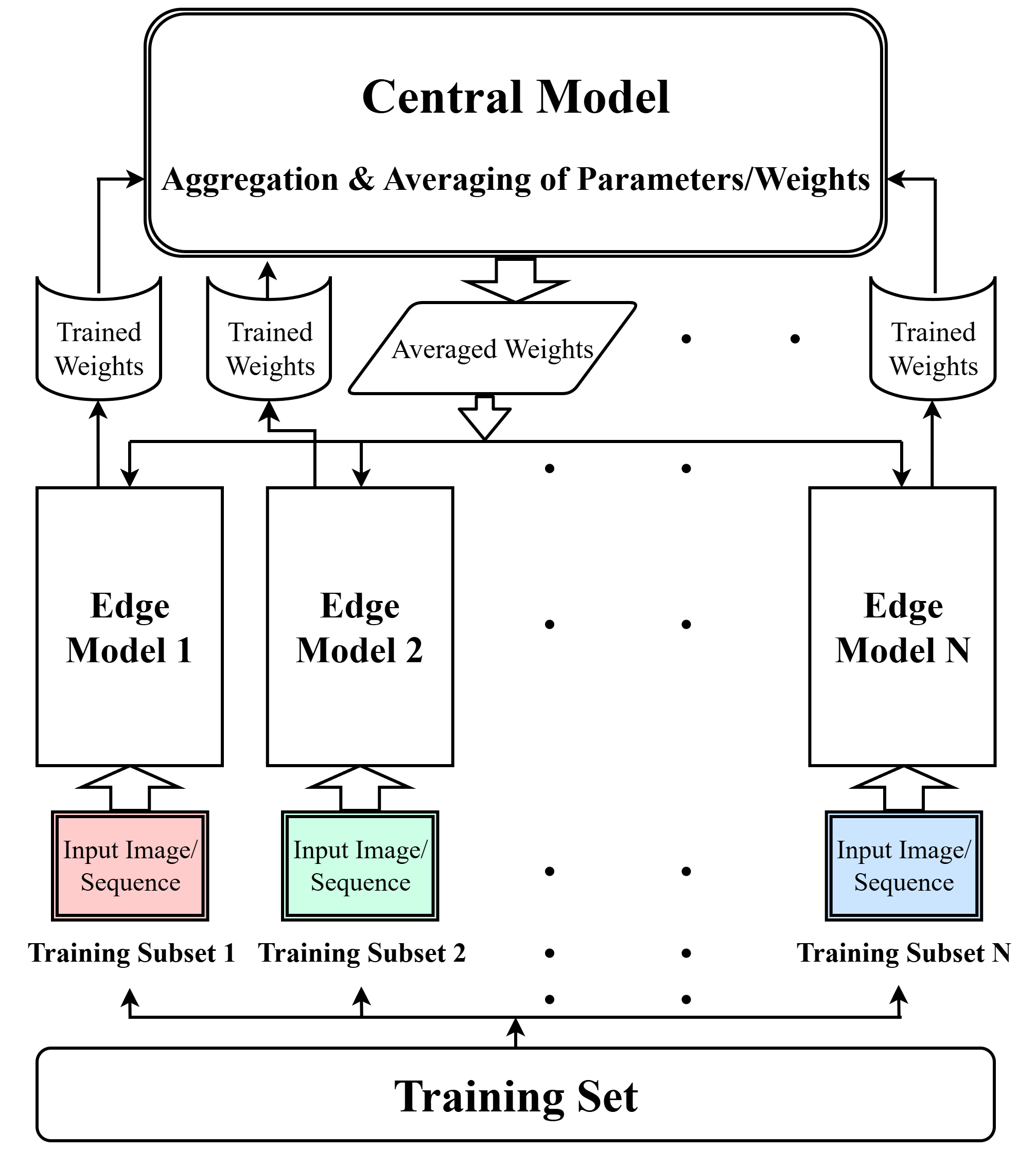
Divide2Conquer (D2C) & Dynamic Uncertainty Aware Divide2Conquer (DUA-D2C): Overfitting Remediation in Deep Learning Using a Decentralized Approach
Supervisor: Prof. Dr. Golam Rabiul Alam
Part 1 (Divide2Conquer): Introduced the Divide2Conquer (D2C) method, which partitions training data to train identical models independently on each subset, aggregating parameters periodically to limit the impact of individual outliers/noise and mitigate overfitting. Published at IEEE BigData 2024.
Part 2 (DUA-D2C): Extended D2C to develop Dynamic Uncertainty-Aware D2C (DUA-D2C), an enhancement that dynamically weights edge models based on performance and prediction uncertainty, further improving robustness. Currently under review.
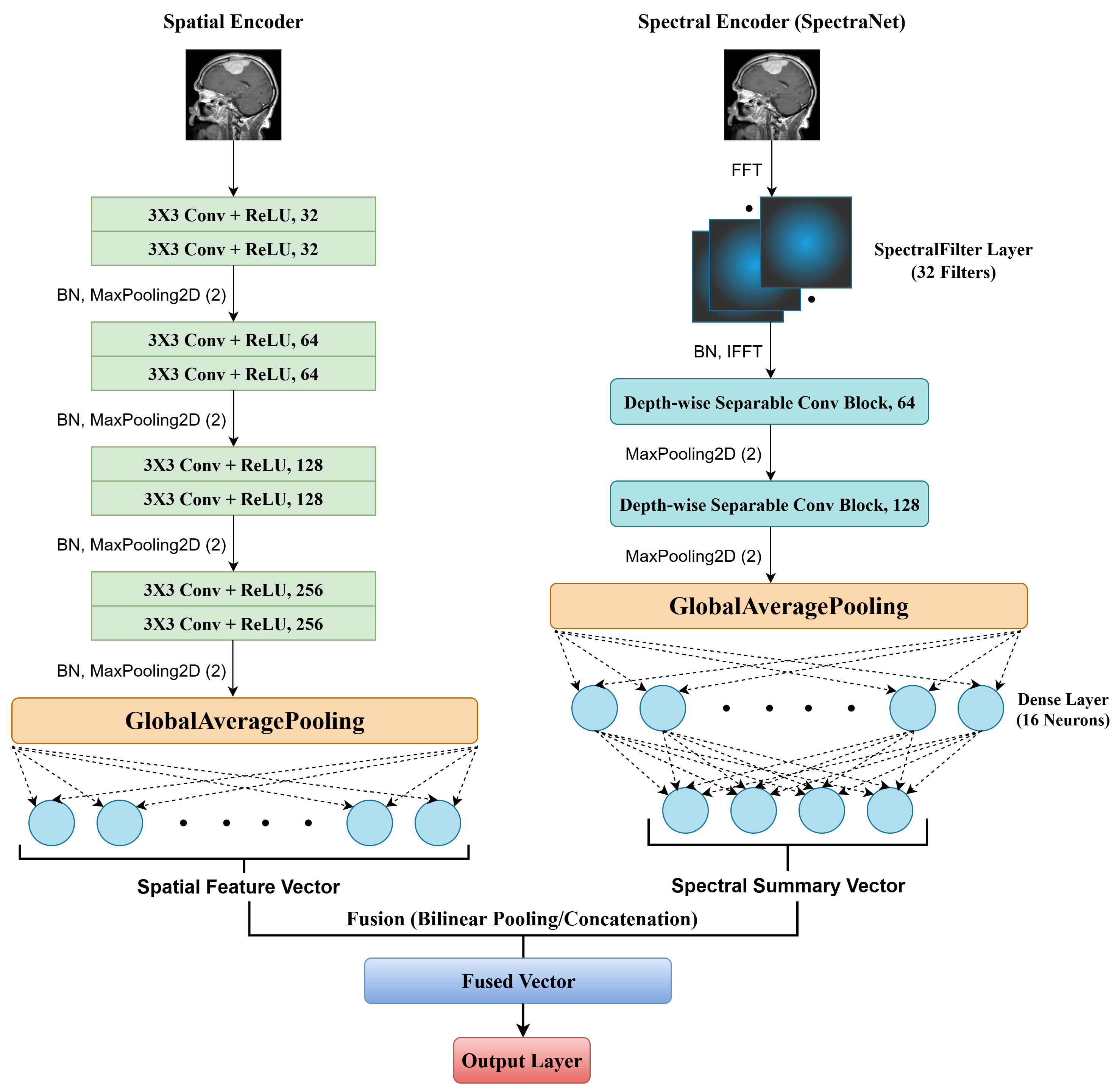
Medical Image Segmentation & Classification based on a Dual-Domain Approach of Spatial and Spectral Feature Fusion
Supervisor: Prof. Dr. Mohammed Imamul Hassan Bhuiyan
Part 1 (S³F-Net): Proposed S³F-Net, a dual-branch framework that learns from both spatial (CNN) and spectral (SpectraNet) domains simultaneously, achieving performance improvements over unimodal baselines & state-of-the-art competitive accuracy on multiple medical imaging datasets.
Part 2 (Spatio-Spectral Medical Image Segmentation): Currently extending the S³F-Net architecture to perform robust and accurate segmentation tasks on medical images, leveraging its multi-domain feature learning capabilities.
Bioradiolocation-Based Multi-Class Sleep Stage Classification
Supervisor: Prof. Dr. Mohammed Imamul Hassan Bhuiyan
This undergraduate thesis focused on a non-invasive sleep stage classification method using bioradiolocation signals. By extensively extracting time & frequency domain features and employing a Random Forest classifier, the system outperformed state-of-the-art methods in terms of several metrics. Published at ICECE 2022.
Ongoing Research Projects
Machine Learning & Statistical Modeling Driven Repression and Mobilization Analysis of Bangladesh's Social Movements
This project applies ML and statistical modeling to analyze the dynamics of state repression and citizen mobilization during Bangladesh's recent social movements. One paper from this work focusing on July Revolution is already under review.
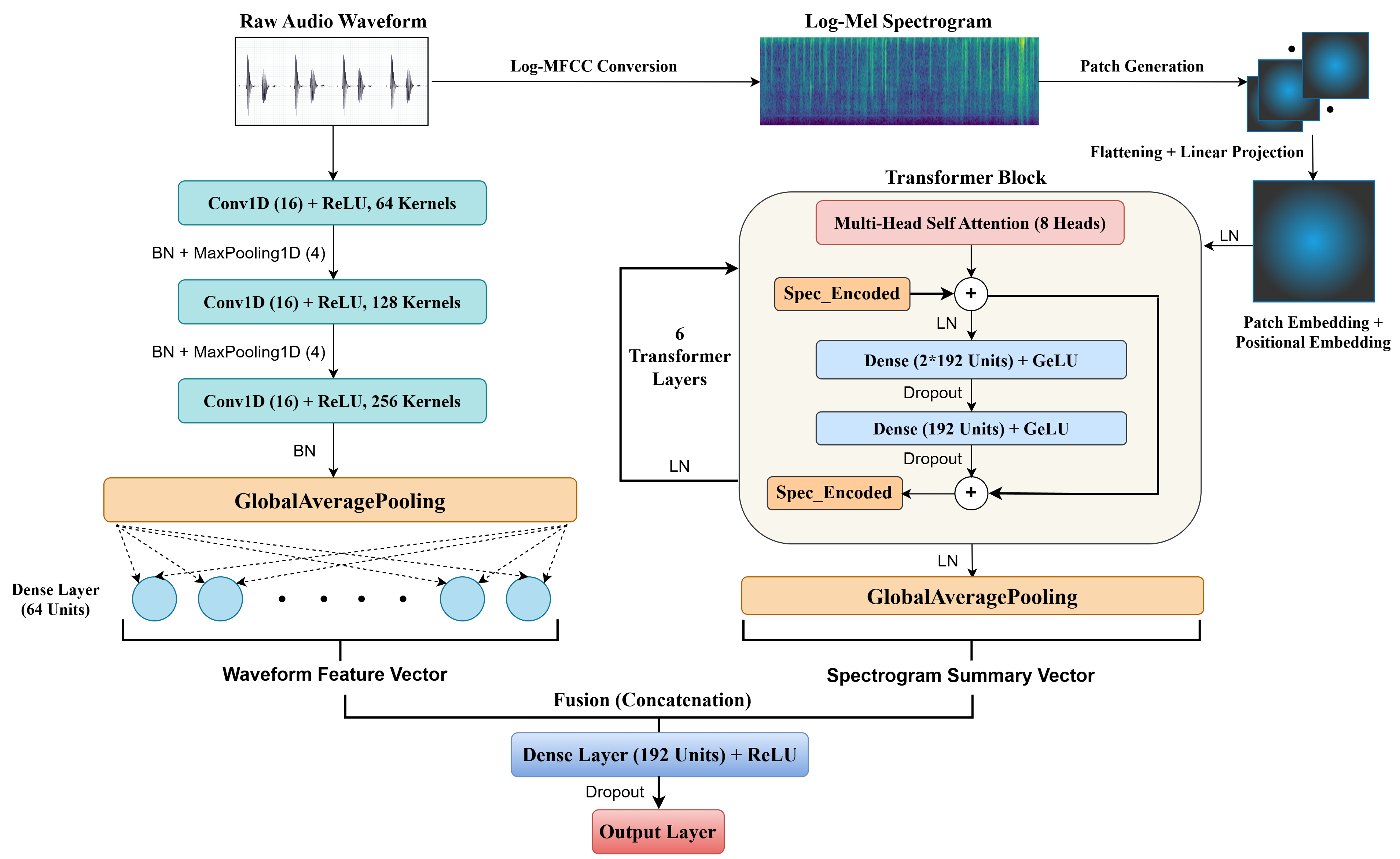
Multi-Modal Fusion of Different Representations of the Signal for Biomedical Audio Analysis
This project explores fusion of features learned from different representations of biomedical audio signals, to exploit the complementary strengths of each representation. We explored different representations including waveforms, spectrograms, and scalograms for robust diagnostic models, and one paper from this research is currently under review.
Antimicrobial Resistance (AMR) Prediction from Genomic Data (SNPs) Leveraging Deep Learning & Explainable AI
This research exploits different perspectives provided by different types of models. We explored the utility of the sequential information available in SNPs through sequence-aware models and currently investigating fusion mechanisms to combine them with "bag-of features" models. We also incorporate Explainable-AI methods to biologically validate our models, by mapping the SNP positions to the corresponding genes and investigating their relevance in AMR. One paper from this research has been accepted at SCA/HPCAsia 2026.
Research Experience
Research Assistant
February - August 2022Center for Computational and Data Sciences (CCDS), IUB
- Investigated ML-based methods of Fetal ECG Separation.
- Contributed to the Graph Neural Networks (GNN) study group.
Co-founder & Researcher
2025 - PresentBIOML (Bioinformatics, Imaging & Omics using Machine Learning) Lab
- Co-founded a collaborative research group of young researchers from Bangladesh.
- Focus on ML applications in bioinformatics, biomedical imaging, biomedical signal processing, and omics.
Research Grants
Co-Principal Investigator
2023 – 2025UIU Research Grant, Institute for Advanced Research (IAR)
- Project Title: “Training Sets vs Training Subsets: Another Method to Reduce Overfitting?”
- Grant Amount: BDT 4,90,000
- Grant ID: UIU-IAR-02-2022-SE-06
Mentorship and Supervision
Supervised or currently supervising 72 students for their undergraduate dissertations, primarily in the fields of Medical Imaging, Bioinformatics, and Health Informatics.
Example Supervised Thesis:
Fusion-Based Multimodal Deep Learning to Improve Detection of Diabetic Retinopathy
Integrating retinal imaging, clinical data and systemic biomarkers to enhance disease detection.
Supervisors: Dr. Jannatun Nur Mukta, Md. Saiful Bari Siddiqui
January '25 - October '25
Thesis ReportConnect
Get in Touch
Find me on academic and professional networks, or reach out via email.